ecic estimates a changes-in-changes model with multiple periods and cohorts as suggested in Athey and Imbens (2006). Changes-in-changes is a generalization of the difference-in-differences approach, estimating a treatment effect for the entire distribution instead of averages.
Athey and Imbens (2006) show how to extend the model to multiple periods and cohorts, analogously to a two-way fixed-effects model for averages. This package implements this, calculating standard errors via bootstrap and plotting results, aggregated or in an event-study-style fashion.
Installation
ecic is available on CRAN using:
install.packages("ecic")You can install the newest version from GitHub:
# install.packages("remotes")
remotes::install_github("frederickluser/ecic")Basic Example
Estimation
Let’s look at a short example how to use the package. First, load some simulated sample data.
library(ecic)
data(dat, package = "ecic")
head(dat)
#> countyreal first.treat year time_to_treat lemp
#> <int> <int> <int> <int> <dbl>
#> 3 1980 1980 0 2.21
#> 3 1980 1981 1 3.33
#> 3 1980 1982 2 3.67
#> 5 1980 1980 0 2.77
#> 5 1980 1981 1 3.88
#> 5 1980 1982 2 3.80Then, the function ecic estimates the changes-changes-model:
# Estimate the model
mod =
ecic(
yvar = lemp, # dependent variable
gvar = first.treat, # group indicator
tvar = year, # time indicator
ivar = countyreal, # unit ID
dat = dat, # dataset
boot = "weighted", # bootstrap proceduce ("no", "normal", or "weighted")
nReps = 10 # number of bootstrap runs
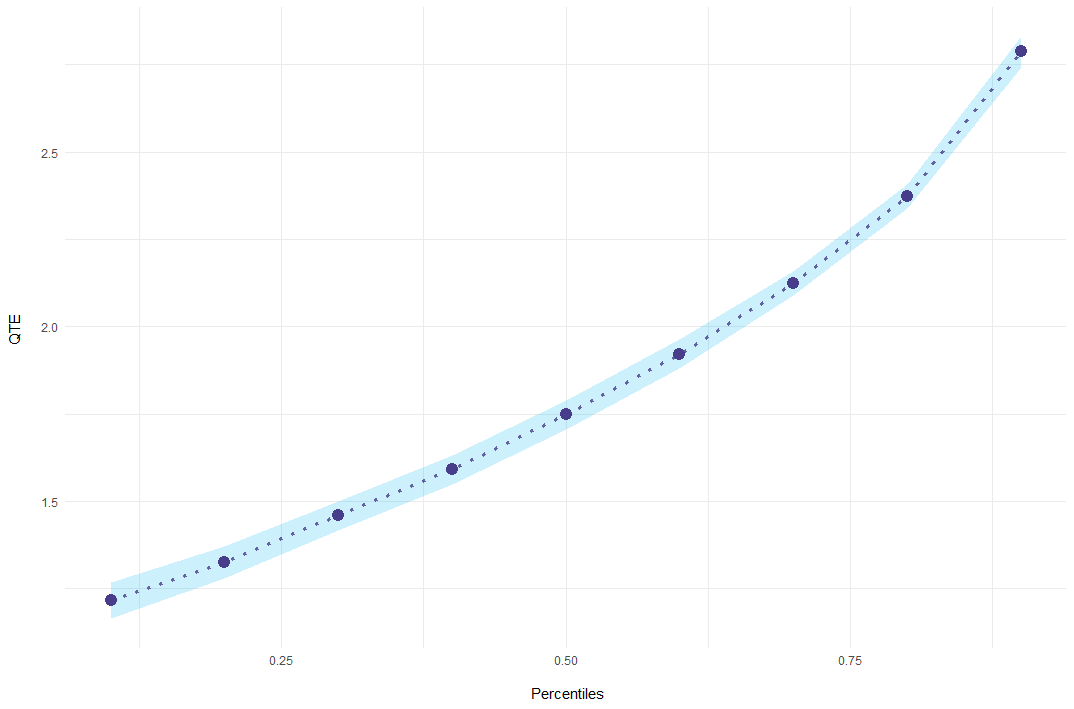
)The input gvar denotes the period in which this individual receives the treatment. mod contains for every bootstrap run the point-estimates. The function summary then combines all bootstrap runs to a quantile treatment effect and adds standard errors:
mod_res = summary(mod)
#> perc coefs se
#> 0.1 1.215531 0.02670761
#> 0.2 1.324130 0.02310521
#> 0.3 1.458270 0.02105119
#> 0.4 1.590848 0.02128534
#> 0.5 1.747296 0.02098057
#> 0.6 1.921818 0.02135982
#> 0.7 2.124138 0.01802972
#> 0.8 2.372483 0.01799869
#> 0.9 2.787395 0.02241811Event-Study Example
The package also allows to report event-study-style results of the effect. To do so, simply add the es = T argument to the estimation and summary will report effects for every event period.
# Estimate the model
mod =
ecic(
yvar = lemp, # dependent variable
gvar = first.treat, # group indicator
tvar = year, # time indicator
ivar = countyreal, # unit ID
dat = dat, # dataset
es = T, # report an event study
boot = "weighted", # bootstrap proceduce ("no", "normal", or "weighted")
nReps = 10 # number of bootstrap runs
)
# report results for every event period
mod_res = summary(mod)
#> [[1]]
#> perc es coefs se
#> 0.1 0 0.9175263 0.02924326
#> 0.2 0 0.9675225 0.02508082
#> 0.3 0 0.9959150 0.02116782
#> 0.4 0 1.0388312 0.02373263
#> 0.5 0 1.0992322 0.02558309
#> 0.6 0 1.1496203 0.03078493
#> 0.7 0 1.2049797 0.03654320
#> 0.8 0 1.2519476 0.03291178
#> 0.9 0 1.3616626 0.01765538
#> [[2]]
#> perc es coefs se
#> 0.1 1 2.393816 0.022273736
#> 0.2 1 2.386941 0.020039276
#> 0.3 1 2.423415 0.017145110
#> 0.4 1 2.452259 0.017982620
#> 0.5 1 2.484616 0.009979006
#> 0.6 1 2.525388 0.012816760
#> 0.7 1 2.575615 0.015196499
#> 0.8 1 2.630959 0.019570320
#> 0.9 1 2.730742 0.024796025
#> [...]Plotting
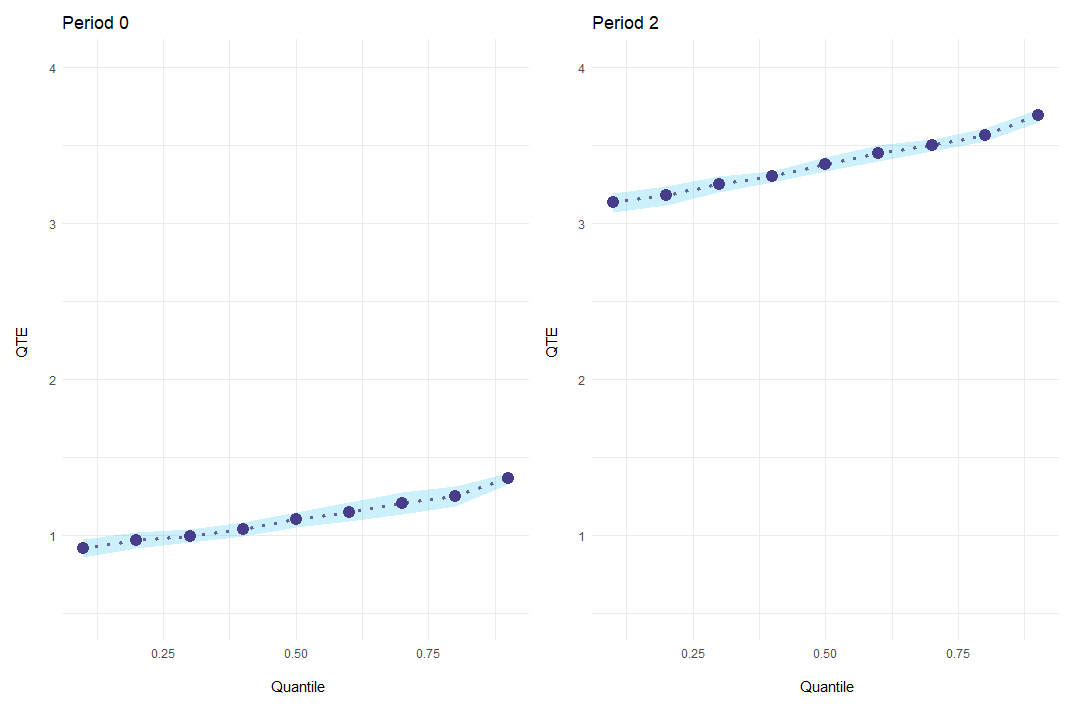
In addition to a standard plot showing everything at once, event-study results can be plotted for every period individually with the option es_type = "for_periods".
ecic_plot(
mod_res,
periods_plot = c(0, 2), # which periods you want to show
es_type = "for_periods", # plots by period
ylim = c(.5, 4) # same y-axis
)
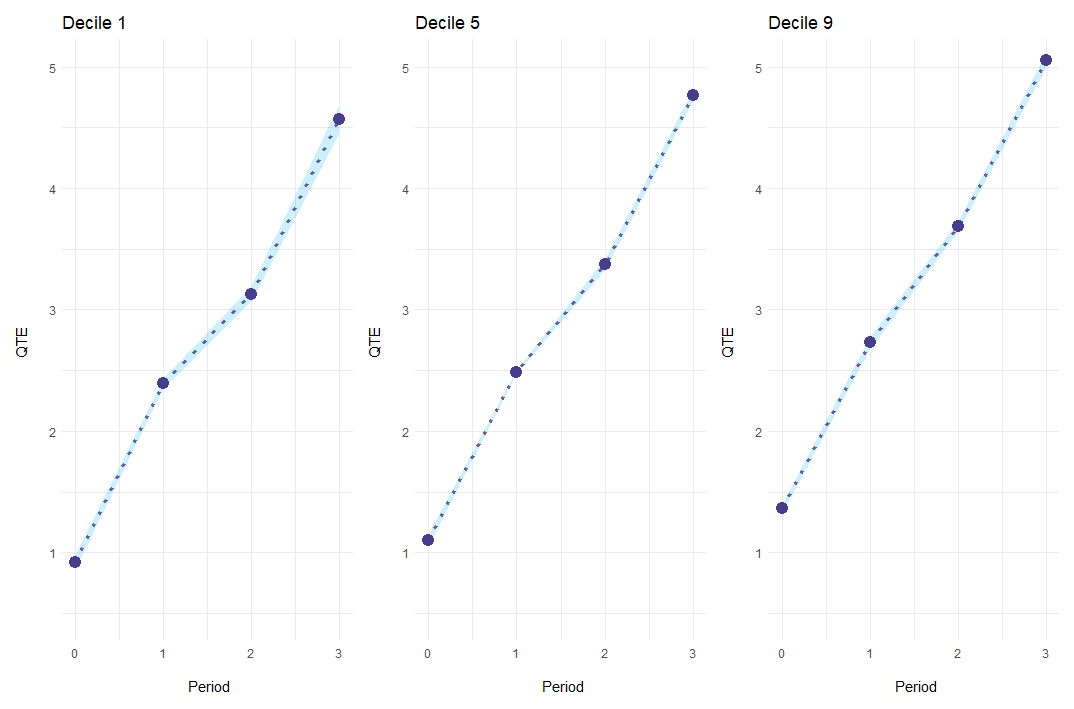
Alternatively, es_type = "for_quantiles" generates one plot for every quantile of interest.
ecic_plot(
mod_res,
periods_plot = c(.1, .5, .9), # which quantiles you want to show
es_type = "for_quantiles", # plots by period
ylim = c(.5, 5) # same y-axis
)
Under the hood
Estimation
For every treated cohort, we observe the distribution of the potential outcome Y(1). In the case of two groups / cohorts and two periods, Athey and Imbens (2006) show how to construct the counterfactual Y(0). This extends to the case with multiple cohorts and periods, where every not-yet-treated cohort is a valid comparison group. Hence, every combination of treated and not-yet-treated cohorts with a common pre-treatment period estimates in theory the quantile treatment effect. Then, I simply want to average them.
Yet, since it is not allowed to simply average quantile treatment effects, we must first store the empirical CDF of Y(1) and Y(0) for every two-by-two case. Next, I aggregate all estimated CDFs to get the plug-in estimates of Y(1) and Y(0), weighting for the cohort sizes. Finally, I invert them to get the quantile function and compute the quantile treatment effect as in the standard case.
Note that it is impossible to estimate a quantile treatment effect for units treated in the first (no pre-treatment period) and last period (no comparison cohort). In addition, the default value of nMin skips small cohorts (default nMin = 40) as we need more observations to estimate quantile treatment effects compared to an average effect.
Speed Improvements
Technically, ecic generates a grid over the dependent variable and imputes all empirical CDFs for every unique value of yvar. You can (cautiously) speed up the imputation by rounding the dependent variable to n_digits.
Bootstrap
I calculate standard errors by bootstrap. I resample with replacement the entire dataset and estimate Y(1) and Y(0) nRep times (default nReps = 1). Bootstrap can either be computed through replacement over the entire dataset (with boot = "normal") or you can weight by cohort sizes (with boot = "weighted") if you worry, for example, about small cohorts. This part can be parallelized by setting nCores > 1, speeding up the computation at the cost of additional overhead to load the cores.
progress_bar prints the progress of the bootstrapping by default. Alternatively, the option progress_bar = "cli" also shows estimated running time, but requires the cli package to be installed.